Biologists are natural collectors. As kids, we jam flowers into heavy books, trap poor bugs in new home-made homes, and curate elaborate collections of sea-shells. As adults, we understand the real purpose of collecting – educating, informing and fascinating.

Photograph by Barta IV.
Today, the systematic collection of biological samples have the potential to provide an incredible resource for genetic studies. By extracting DNA from old specimens, including feathers, bones and hair, we could examine a range of subjects, such as:
- How populations have changed over time;
- The genetic consequences of population collapses and inbreeding;
- The impact of interbreeding with non-native individuals.
However, such studies in historical samples have had a reputation for being problematic and unreliable, and for good reason. Over long periods of time, DNA starts to degrade and fragment into smaller pieces, making it harder to screen genetic markers in older samples. In addition, old samples are more open to contamination with related DNA, making the results less reliable.
Fortunately, new genotyping technologies can examine single nucleotide polymorphisms (genetic markers known as SNPs) in much smaller fragments of DNA. But how reliable are these results? Can we really use old samples for cutting edge genetic studies?
During my time with the Primmer Group at the University of Turku, I was interested in understanding the evolution of a number of life-history traits in Atlantic salmon in the Teno river system in Northern Finland. But, getting up to such a remote place and collecting enough salmon fitting our study design was just not possible. Instead, we decided to use a pre-existing resource – a huge collection of thousands of phenotypic measurements and scale samples by the fishermen on Teno river over the last 40 years, curated by the Finnish Game and Fisheries Research Institute in Utsjoki.
We were excited by the prospect of using new SNP technology in our old fish scales. Initial pilot genotyping looked very promising, but there was the still the worry that somehow, our results could be seen as unreliable. Therefore, we decided to put this idea to rest by formally testing how efficient and repeatable SNP genotyping was.
We set about examining the DNA concentration and fragment sizes in scale samples collected over a 31 year period, repeating DNA extractions and genotyping runs multiple times within the same individual. We found that although DNA concentration didn’t change over time, DNA was much more fragmented in older samples. Despite this, the genotyping method we used (an Illumina iSelect SNP array) gave us more than 97% genotyping success and and error rate of < 0.2% when scanning more than 4,000 SNP markers in samples up to 35 years old.

Temporal variation of DNA quality in the Teno scale samples in 48 genotyping runs in 16 fish. A. Sample call rate and year. B. Sample call rate in relation to the proportion of DNA with a fragment size of > 1000bp. [NB. Samples in red come from a single fish which had a particularly low call rate and higher error rate. This sample would not have been included in any further genetic study.]
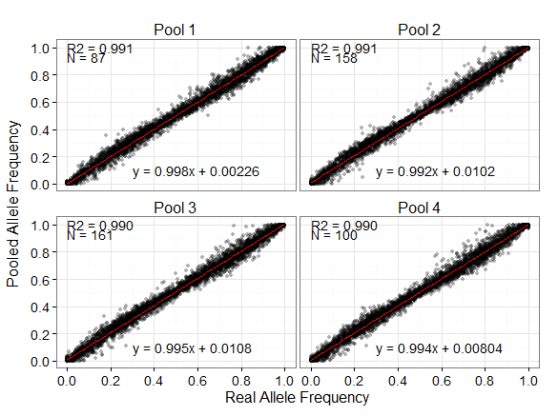
Correlation between pooled allele frequencies and real allele frequencies within each pool. Pooled allele frequencies shown here were calculated from nine replicates within each pool. R2 is the adjusted R2 values from a linear regression. N is the number of individuals included in each pool.
Overall, our findings gave us confidence that more detailed genetic studies are now much more achievable in old DNA samples as a result of SNP genotyping technologies. This is not just in Atlantic salmon – these methods are applicable to many different species. We have published our findings and conclusions in the journal BMC Genomics, along with detailed methods, results and recommendations for future studies.
Johnston SE, M Lindqvist, E Niemelä, P Orell, J Erkinaro, MP Kent, S Lien, J-P Vähä, A Vasemägi, CR Primmer (2013) Fish scales and SNP chips: SNP genotyping and allele frequency estimation in individual and pooled DNA from historical samples of Atlantic salmon (Salmo salar). BMC Genomics, 14:439. [Open Access Link]

